Neanderthal Man (2 page)

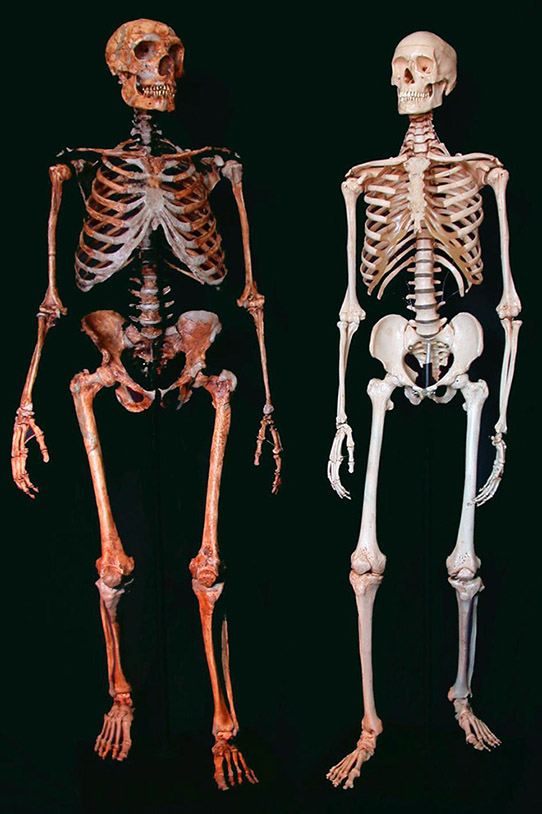
Figure 1.1. A reconstructed Neanderthal skeleton (left) and a present-day human skeleton (right). Credit: Ken Mowbray, Blaine Maley, Ian Tattersall, Gary Sawyer, American Museum of Natural History.
As interesting as these questions were in themselves, it seemed to me that the Neanderthal bone fragment held the promise of an even larger prize. Neanderthals are the closest extinct relative of contemporary humans. If we could study their DNA, we would undoubtedly find that their genes were very similar to ours. Some years earlier, my group had sequenced a large number of DNA fragments from the chimpanzee genome and had shown that in DNA sequences we shared with the chimpanzees, only a bit over 1 percent of the nucleotides differed. Clearly, the Neanderthals must be much closer to us than that. But—and this is what was immensely exciting—among the few differences one would expect to find in the Neanderthal genome, there must be those that set us apart from all earlier forms of human forerunners: not just from the Neanderthals but also from Turkana Boy, who lived some 1.6 million years ago; Lucy, some 3.2 million years ago; and Peking Man, more than half a million years ago. Those few differences must form the biological foundations of the radically new direction our lineage took with the emergence of modern humans: the advent of rapidly developing technology, of art in a form we today immediately recognize as art, and maybe of language and culture as we now know it. If we could study Neanderthal DNA, all this would be within our grasp. Wrapped in such dreams (or delusions of grandeur), I finally drifted off to sleep as the sun rose.
The next day Matthias and I both arrived late at the lab. After checking the DNA sequence from the night before to make sure we had not made any mistakes, we sat down and planned what to do next. It was one thing to get the sequence of one little piece of mtDNA that looked interesting from the Neanderthal fossil, but it would be quite another to convince ourselves, let alone the rest of the world, that it was mtDNA from an individual who lived (in this particular case) some 40,000 years ago. My own work over the previous twelve years made our next step fairly clear. First, we needed to repeat the experiment—not just the last step but all the steps, beginning with a new piece of the bone in order to show that the sequence we had obtained was not some fluke derived from a badly damaged and modified modern mtDNA molecule in the bone. Second, we needed to extend the sequence of mtDNA we had obtained by retrieving overlapping DNA fragments from the bone extract. This would enable us to reconstruct a longer mtDNA sequence, with which we could begin to estimate just how different the mtDNA of Neanderthals was from that of humans today. And then a third step was necessary. I myself had often suggested that extraordinary claims about DNA sequences from ancient bones require extraordinary evidence—namely, repetition of the results in another lab, an unusual step in a typically competitive scientific field. The claim that we had retrieved Neanderthal DNA would certainly be considered extraordinary. To exclude unknown sources of error in our lab, we needed to share some of the precious bone material with an independent lab and hope that it could manage to repeat our result. All of this I discussed with Matthias and Ralf. We laid out plans for the work and swore one another to absolute secrecy outside our research groups. We wanted no attention until we were sure that what we had was the real thing.
Matthias got to work at once. Having spent almost three years on mostly fruitless attempts to extract DNA from Egyptian mummies, he was energized by the prospect of success. Ralf seemed frustrated over having to return to Bonn, where he could do nothing but eagerly await word of our results. I tried to concentrate on my other projects, but it was hard to take my mind off what Matthias was doing.
What Matthias needed to do was not all that easy. We were dealing, after all, with something other than the intact and pristine DNA that comes from a blood sample drawn from a living person. The neat and tidy double-stranded, helical DNA molecule in the textbooks—with its nucleotides A, T, G, and C, attached in complementary pairs (adenine with thymine, guanine with cytosine) to the two sugar-phosphate backbones—is not a static chemical structure when stored in the nuclei and mitochondria of our cells. Rather, DNA continually suffers chemical damage, which is recognized and repaired by intricate mechanisms. In addition, DNA molecules are extremely long. Each of the twenty-three pairs of chromosomes in the nucleus comprises one enormous DNA molecule; the total length of one set of twenty-three chromosomes adds up to about 3.2 billion nucleotide pairs. Since the nucleus has two copies of the genome (each copy stored on one set of twenty-three chromosomes, of which we inherit one from our mother and one from our father), it contains about 6.4 billion nucleotide pairs. By comparison, the mitochondrial DNA is tiny, with a little over 16,500 nucleotide pairs; but given that the mtDNA we had was ancient, the challenge involved in sequencing it was great.
The most common type of damage that occurs spontaneously in DNA molecules, whether nuclear DNA or mtDNA, is the loss of a chemical component—an amino group—from the cytosine nucleotide (C), turning it into a nucleotide that does not naturally occur in DNA called uracil, abbreviated U. There are enzyme systems in the cells that remove these U’s and replace them with the correct nucleotide, C. The discarded U’s end up as cellular garbage, and from analyses of damaged nucleotides excreted in our urine it has been calculated that about ten thousand C’s per cell morph into U’s each day, only to be removed and then replaced. And this is just one of several types of chemical assaults our genome suffers. For example, nucleotides are lost, creating empty sites that quickly lead to breakage of the strands in the DNA molecules. Working against this are enzymes that fill in such missing nucleotides before a break can occur. If a break does occur, other enzymes join the DNA molecules back together. In fact, the genomes in our cells would not remain intact for even an hour if these repair systems were not there to maintain them.
These repair systems, of course, require energy to work. When we die, we stop breathing; the cells in our body then run out of oxygen, and as a consequence their energy runs out. This stops the repair of DNA, and various sorts of damage rapidly accumulate. In addition to the spontaneous chemical damage that continually occurs in living cells, there are forms of damage that occur after death, once the cells start to decompose. One of the crucial functions of living cells is to maintain compartments where enzymes and other substances are kept separate from one another. Some of these compartments contain enzymes that can cut DNA strands and are necessary for certain types of repair. Other compartments contain enzymes that break down DNA from various microorganisms that the cell may encounter and engulf. Once an organism dies and runs out of energy, the compartment membranes deteriorate, and these enzymes leak out and begin degrading DNA in an uncontrolled way. Within hours and sometimes days after death, the DNA strands in our body are cut into smaller and smaller pieces, while various other forms of damage accumulate. At the same time, bacteria that live in our intestines and lungs start growing uncontrollably when our body fails to maintain the barriers that normally contain them. Together these processes will eventually dissolve the genetic information stored in our DNA—the information that once allowed our body to form, be maintained, and function. When that process is complete, the last trace of our biological uniqueness is gone. In a sense, our physical death is then complete.
However, nearly each one of the trillions of cells in our body contains the entire complement of our DNA. Thus, even if the DNA in some cells in some remote corner of our body evades complete decomposition, some trace of our genetic makeup will survive. For example, the enzymatic processes that degrade and modify the DNA require water to work. If some parts of our body dry out before the degradation of the DNA has run its course, these processes will stop, and fragments of our DNA may survive for a longer time. This occurs, for example, when a body is deposited in a dry place where it becomes mummified. Such whole-body desiccation may occur accidentally, owing to the environment in which the body happens to end up, or it may be deliberately practiced. As is commonly known, ritual mummification of the dead was often performed in ancient Egypt, where the bodies of hundreds of thousands of people who lived between about 5,000 and 1,500 years ago were mummified to provide
post mortem
abodes for their souls.
Even when mummification does not occur, some parts of the body, such as the bones and teeth, may survive long after burial. These hard tissues contain cells, responsible for tasks such as forming new bone when a bone is broken, that are embedded in microscopically small holes. When these bone cells die, their DNA may leak out and become bound to the mineral component of the bone, where it may be shielded from further enzymatic attack. Thus, with luck, some DNA may survive the onslaught of degradation and damage that occurs in the immediate aftermath of death.
But even when some DNA survives the bodily chaos that follows death, other processes will continue to degrade our genetic information, albeit at a slower rate. For example, the continuous flow of background radiation that hits Earth from space creates reactive molecules that modify and break DNA. Furthermore, processes that require water—such as the loss of amino groups from the nucleotide C, resulting in U nucleotides—continue even when DNA is preserved under relatively dry conditions. This is because DNA has such an affinity for water that even in dry environments water molecules are almost always bound to the grooves between the two DNA strands, allowing spontaneous water-dependent chemical reactions to occur. The loss of the amino group—or deamination—of the nucleotide C is one of the fastest of these processes, and it will destabilize the DNA so that its strands eventually break. These and other processes, most still unknown, keep chipping away at our DNA even when it has survived the havoc that death itself causes in our cells. Although the rate of despoliation will depend on many circumstances such as temperature, acidity, and more, it is clear that even under favorable conditions, ultimately even the last pieces of information from the genetic program that made a person possible will eventually be destroyed. It seemed that in the Neanderthall bone my colleagues and I had analyzed, all these processes had not yet fully completed their destructive task after 40,000 years.
Matthias had retrieved the sequence of a piece of mtDNA 61 nucleotides long. To do so, he had to make many copies of this piece of DNA, which involved a process called the polymerase chain reaction (PCR). In his attempt to confirm our findings, he started by repeating his PCR experiment exactly as he had done it the first time. This experiment involves using two short synthetic pieces of DNA called primers, which were designed to bind to two places in the mtDNA, 61 nucleotide pairs apart. These primers are mixed with a tiny amount of the DNA extracted from the bone and an enzyme called DNA polymerase that can synthesize new DNA strands starting and ending with the primers. The mixture is heated to allow the two DNA strands to come apart, so that the primers can find and bind to their target sequences when the mixture is cooled via pairings of A’s with T’s and G’s with C’s. The enzyme will then use the primers bound to the DNA strands as starting points to synthesize two new strands, duplicating the two original strands from the bone, so that these two original strands become four strands. This amplification process is repeated to produce eight strands, then again to produce sixteen, then thirty-two, and so on for a total of thirty or forty rounds of duplication.
The PCR—a simple yet elegant technique invented by the maverick scientist Kary Mullis in 1983—is extremely powerful. From a single DNA fragment, one can, in principle, obtain about a trillion copies after forty cycles. This is what made our work possible, so in my opinion Mullis certainly deserved the Nobel Prize in chemistry that he was awarded in 1993. However, the PCR’s exquisite sensitivity also made our work difficult. In an extract from an ancient bone—which may contain very few surviving ancient DNA molecules, or none at all—there might be one or more molecules of modern human DNA that have contaminated the experiment: from the chemicals used, from the lab’s plasticware, or from airborne dust. Dust particles in rooms where humans live or work are, to a large extent, human skin fragments, which contain cells full of DNA. Alternatively, human DNA might have contaminated the sample when a person handled it—say, in a museum or at an excavation. It was with these concerns in mind that we chose to study the sequence of the most variable part of the Neanderthal mtDNA. Since many humans differ from one another in that particular section, we could at least tell whether more than one human had contributed DNA to our experiment and thus be warned that something was amiss. This is why we were so excited about finding a DNA sequence with changes never before seen in any human; had the sequence looked like that of a living human, we would have had no way of determining whether it meant, on the one hand, that the Neanderthal was indeed identical in mtDNA to some people today or, on the other, that we were just looking at a modern mtDNA fragment that had made its way into our experiments from some insidious source such as a speck of dust.
