Life on a Young Planet (5 page)
Read Life on a Young Planet Online
Authors: Andrew H. Knoll

The cyanobacteria comprise only one of five distinct groups of photosynthetic bacteria. In the other groups, electron supply by H
2
S, hydrogen gas (H
2
), or organic molecules is obligatory, and oxygen is never produced. These photosynthetic bacteria harvest light using bacteriochlorophyll rather than the more familiar chlorophyll. Some employ the same biochemistry as cyanobacteria and green plants to fix carbon dioxide, but others have distinctively different pathways, and still others rely on carbon already packaged into organic molecules.
Bacterial variations on the metabolic themes of respiration, fermentation, and photosynthesis are, thus, impressive, but prokaryotic organisms have evolved yet another way of growing that is completely unknown in eukaryotes:
chemosynthesis
. Like photosynthetic organisms, chemosynthetic microbes get their carbon from CO
2
, but they harvest energy from chemical reactions rather than sunlight. Oxygen or nitrate (or, less commonly, sulfate, oxidized iron, or manganese) is combined with hydrogen gas, methane, or reduced forms of iron, sulfur, and nitrogen in ways that allow the cell to capture the energy released by the reaction. Methanogenic prokaryotes are of particular evolutionary and ecological interest; these tiny cells can gain energy from the reaction of hydrogen gas and carbon dioxide to produce methane.
The metabolic pathways of prokaryotes sustain the chemical cycles that maintain Earth as a habitable planet. Take carbon dioxide, for example. Volcanoes supply CO
2
to the oceans and atmosphere, but photosynthesis removes it at a far faster clip. So much faster, in fact, that photosynthetic organisms could strip the present-day atmosphere of its CO
2
in little more than a decade. They don’t, of course, principally because respiration, in essence, runs the photosynthetic reaction backward. While photosynthetic organisms react CO
2
and water to produce sugar and oxygen, respiring creatures (including you, as you read this sentence) react sugar with oxygen, giving off water and carbon dioxide. Together, photosynthesis and respiration
cycle
carbon through the biosphere, sustaining life and maintaining the environment through time.
It is easy to envision a simple carbon cycle in which cyanobacteria fix CO
2
into organic matter and supply oxygen to the environment, while respiring bacteria do the reverse, consuming oxygen and regenerating CO
2
. Plants and algae would do just as well as cyanobacteria, and protozoa, fungi, and animals could substitute for bacterial respirers—the prokaryotes and eukaryotes are functionally equivalent. But let’s let some cells sink to the seafloor and become buried in oxygen-depleted sediments. Now the limitations of eukaryotic metabolism become clear—reactions that do not use oxygen (
anaerobic
reactions) are needed to complete the carbon cycle. In modern seafloor sediments, sulfate reduction and respiration using iron and manganese are just as important as aerobic respiration in recycling organic matter. More generally, wherever carbon passes through oxygen-free environments, bacteria are essential to the carbon cycle; eukaryotes are everywhere optional.
The fundamental importance of prokaryotes extends to other biologically important elements, as well. Indeed, in the biogeochemical cycles of sulfur and nitrogen,
all
the principal metabolic pathways that cycle these elements are prokaryotic. Consider, in particular, nitrogen, an essential element required for the formation of proteins, nucleic acids, and other biological compounds. We live our lives bathed in nitrogen gas. (Air is about 80 percent N
2
by volume.) But this vast repository of nitrogen is not biologically available to us; like other animals, we obtain the nitrogen we need by eating other organisms. As it turns out, nitrogen gas is no more available to cattle or corn than it is to humans. Plants can take up ammonium (NH
4
+
) or nitrate from the soil, but how do these compounds get there in the first place? Ammonium is released as dead cells decay; nitrate, in turn, is produced by bacteria that oxidize ammonium. In oxygen-rich habitats, the resulting nitrate is available to plants (or, in aquatic ecosystems, algae and cyanobacteria), but in waterlogged soil or other environments where O
2
is depleted, other bacteria use nitrate for respiration, returning nitrogen to the atmospheric pool of N
2
. (Much of the nitrate spread across fields as fertilizer is lost in this way.)
So, we haven’t solved our problem. The ammonium and nitrate in soil and seawater come from dead cells, and nitrate-respiring bacteria inexorably remove biologically usable nitrogen from the environment. What, then, fuels the biological nitrogen cycle and keeps it from running down?
The answer is that some organisms are able to convert atmospheric nitrogen to ammonium, using the cell’s store of energy.
No
eukaryotic organism can fix nitrogen in this way, but many prokaryotes can. (Farmers commonly include soy or other beans in their crop rotation because these plants restore nitrogen to the soil. The task of nitrogen fixation, however, is accomplished by bacteria that live in small nodules on the roots of bean plants, not by the beans themselves.) A small amount of nitrogen is fixed by lightning as it cuts through the atmosphere, but biology’s thirst for nitrogen is quenched mainly by bacteria.
The cycles of carbon, nitrogen, sulfur, and other elements are linked together into a complex system that controls the biological pulse of the planet. Because organisms need nitrogen for proteins and other molecules, there could be no carbon cycle without nitrogen fixation. Nitrogen metabolism itself depends on enzymes that contain iron; thus, without biologically available iron, there could be no nitrogen cycle … and, hence, no carbon cycle. Biology on another planet may or may not include organisms that are large or intelligent, but wherever it persists for long periods of time, life will feature complementary metabolisms that cycle biologically important elements through the biosphere.
By now it should be apparent why I insisted earlier that plants and animals evolved to fit into a prokaryotic world rather than the reverse. It
is
a prokaryotic world, and not only in the trivial sense that there are a lot of bacterial cells. Prokaryotic metabolisms form the fundamental ecological circuitry of life. Bacteria, not mammals, underpin the efficient and long-term functioning of the biosphere.
How can this astonishing diversity of prokaryotic cells be ordered and assembled along with that of eukaryotes into a phylogeny that encompasses all of biology? Size and shape fail us, and so does physiology; organisms as disparate as fungi and elephants, or
E. coli
and redwoods, are simply too different from one another to assemble into a believable tree based on form and function alone. The solution requires that we return to the unity of life, the molecular attributes shared by all known organisms. In a groundbreaking paper published in 1965, Emile Zuckerkandl and Nobel laureate Linus Pauling proposed that molecules can be read as documents of evolutionary history. Just as the anatomical
structures of limbs or skulls reflect descent with modification, so too do the chemical structures of DNA and proteins. The long chain of amino acids that makes up, say, the respiratory protein cytochrome c differs slightly between humans and chimps and more so between humans/chimps and horses. The sequences of nucleotides in the genes that code for these proteins differ correspondingly.
Carl Woese of the University of Illinois built decisively on this conceptual foundation. Woese spent his formative years in science investigating ribosomes, the sites within cells where proteins are manufactured. He knew that all organisms contain ribosomes, that all ribosomes contain functional complexes made of RNA and proteins, and that these complexes all contain several subunits. By comparing among organisms the sequences of nucleotides that make up the RNA molecules found in the small subunit of ribosomes, Woese made the great leap that brought phylogeny to the microbial world, sowing the seeds for a Tree of Life worthy of the name.
Figure 2.1
shows the Tree of Life, a depiction of the genealogical relationships of all living organisms, based on comparisons of molecular sequence in the genes that code for small subunit ribosomal RNA. Experts argue about its details, but all biologists agree that our ability to draw Darwin’s great Tree of Life in its entirety constitutes one of the great intellectual achievements of the late twentieth century.
The first thing to notice about the tree is that it contains three major limbs, termed
domains
by Woese. Two of the domains are unsurprising: the eukaryotes and the bacteria fall on distinct branches. The third, however, came as a shock when Woese and then postdoctoral fellow George Fox proposed it in 1977. The Archaea are prokaryotic in cell organization, and for many years the organisms on this branch had been thought of (when they were thought about at all) as metabolically unusual bacteria. But comparison of ribosomal RNA genes suggests that these microbes are fully as distinct from the conventional bacteria as bacteria are from eukaryotes. What’s more, the tree indicates that archaeans are actually more closely related to the eukaryotes than they are to bacteria. (In phylogenetic discourse, closeness of relationship reflects recency of common ancestry; it is a statement about genealogy, not similarity.)
The complete genome (genetic information encoded in DNA) of the archaean
Methanococcus janaschii
was published in 1996, revealing that this microbe shares just 11–17 percent of its genes with bacteria whose genomes have been sequenced. More than 50 percent of its genes are unknown in
either
eukaryotes or bacteria, confirming that archaeans are distinctly different from organisms in the other two domains. Archaeans do, however, have some important characters in common with bacteria, such as (most obviously) prokaryotic cell organization, the molecular structure of the ribosome, and the arrangement of genes on a single circular chromosome. Equally, on the other hand, archaeans share attributes such as molecular details of DNA transcription and susceptibility to specific antibiotics with eukaryotes. And there are still other traits that
bacteria and eukaryotes share to the exclusion of Archaea—prominent among these is the nature of the cell membrane.
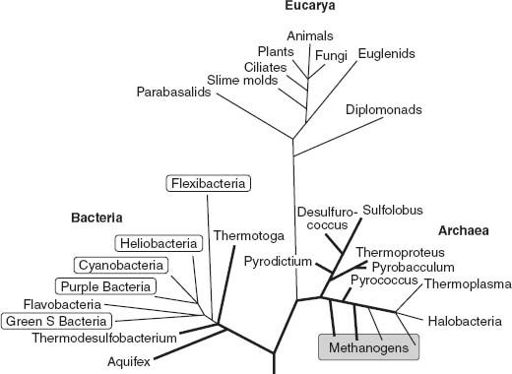
Figure 2.1.
The Tree of Life, a depiction of the genealogical relationships of living organisms, based on sequence comparisons of genes that code for RNA in the small subunit of the ribosomes found in all cells. Note the three principal branches, made up of Bacteria, Archaea, and Eucarya. Branch lengths indicate degree of difference among gene sequences; because genes can evolve at different rates, however, this does not necessarily translate into time. Bacterial groups with photosynthetic members are highlighted by clear boxes; methanogenic archeans fall within the shaded box. Heavy lines denote hyperthermophiles—groups of organisms that live at high temperatures. (Adapted from a depiction of the tree by Karl Stetter)
How then do we tell who is more closely related to whom? Put another way, where do we place the
root
on this tree? A three-branched tree can’t be rooted by conventional means, and a bit more consideration of character distributions shows why. Features such as ATP or the genetic code that are shared by all three domains carry no information on genealogical relationships, but permit inferences about the nature of the last common ancestor of the three branches. In contrast, attributes such as cell wall composition that are distinct in each limb provide no information on either genealogy or ancestral features. Characters shared by two of the three domains would appear to offer better prospects for tree building, but such distributions can be explained equally well in several different ways. For example, if we assume that membranes composed of fatty acids were present in the last common ancestor, then we can posit that this trait was retained in bacteria and eukaryotes but replaced by isoprenoid-based membranes along the road to the Archaea. Alternatively, we can assume that membranes built from isoprenoids are ancestral, but were swapped for fatty-acid membranes in the common ancestor of bacteria and eukaryotes. Like the first alternative, this tree requires only one evolutionary change. (We could, of course, eliminate some possibilities if we knew for certain which traits characterized the last common ancestor, but we have no way of determining this.)
A clever solution to the rooting problem was proposed in 1989. Two research groups headed by Naoyuki Iwabe and Peter Gogarten, respectively, independently recognized that while three sets of organisms cannot be joined into a rooted tree, some of the genes they contain can be. The genes in question share a specific property: they were present in duplicate sets in the last common ancestor. How does this help us? As shown in
figure 2.2
, each of the two sister genes present in the last common ancestor diverged as the three domains differentiated. The resulting array of genes can be ordered into a tree. In toto, the tree is unrooted, but it consists of two component branches that can be rooted
relative
to each other. Both “half trees” have the same form: one branch that contains only bacteria and a second containing Archaea plus eukaryotes.
